Experiments III: Cell-Free Fluorescence
Source:vignettes/expts_iii_cell_free.Rmd
expts_iii_cell_free.Rmd
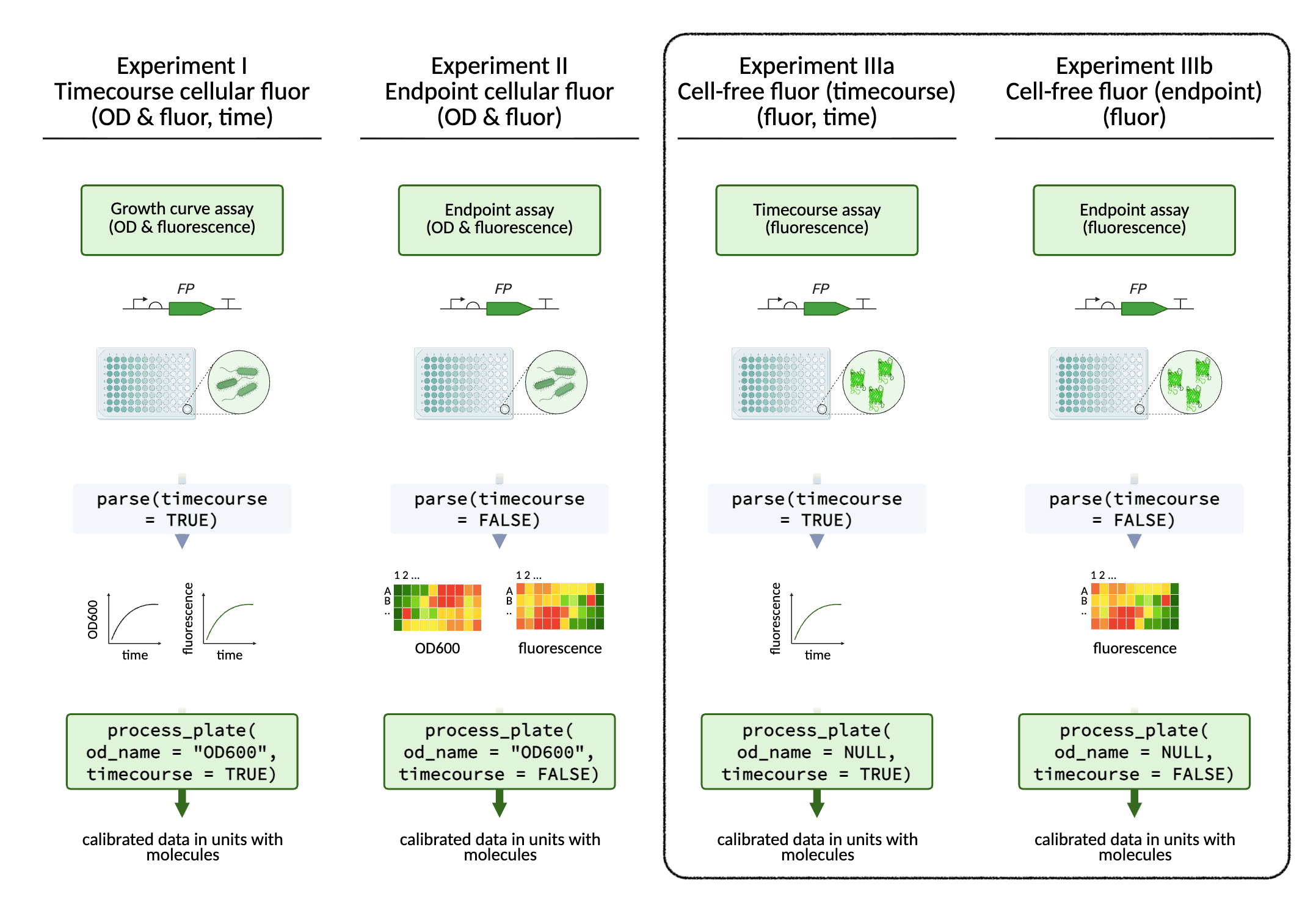
In addition to cellular fluorescence data, cell-free fluorescence
data can also be processed with FPCountR, using the same functions. Just
apply the argument od_name = NULL.
Processing data from cell-free fluorescent protein experiments
Example for cell-free data
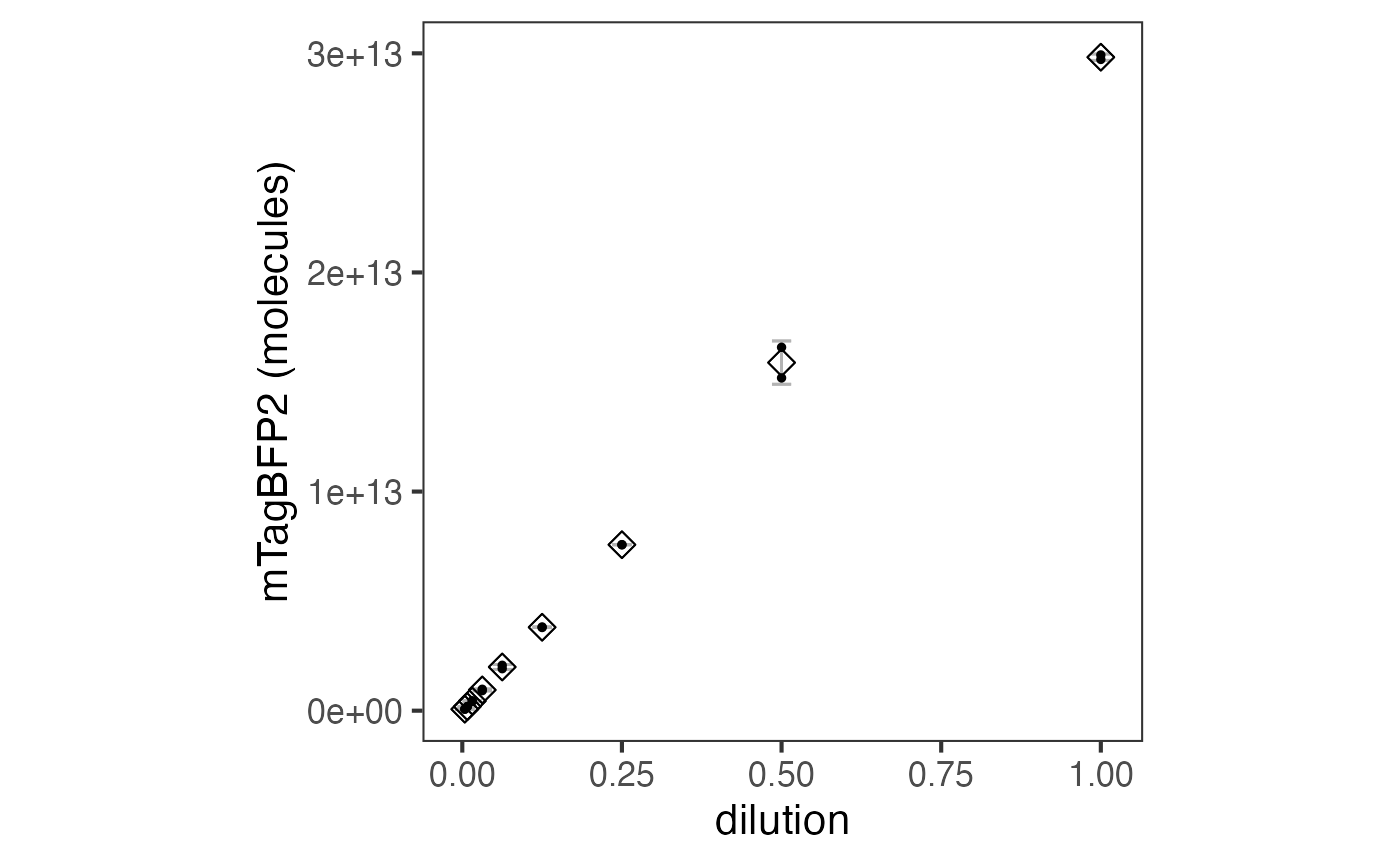
Let’s consider an example in which cell-free fluorescence was measured in vitro. For this we will use a dilution series of mTagBFP2, which was done with purified protein. This data has already been parsed. With conversion factors for calibrating mTagBFP2 fluorescence in hand, we are ready to process the experimental data.
parsed_data <- read.csv("data/example_fluorescence_simple_parsed.csv")
parsed_data[1:24,] # view a fragment of the dataframe| protein | replicate | volume | dilution | well | blueblue090 | row | column |
|---|---|---|---|---|---|---|---|
| mTagBFP2 | 1 | 200 | 1.000000000 | A1 | 50522 | A | 1 |
| mTagBFP2 | 1 | 200 | 0.500000000 | A2 | 27526 | A | 2 |
| mTagBFP2 | 1 | 200 | 0.250000000 | A3 | 15485 | A | 3 |
| mTagBFP2 | 1 | 200 | 0.125000000 | A4 | 9504 | A | 4 |
| mTagBFP2 | 1 | 200 | 0.062500000 | A5 | 6530 | A | 5 |
| mTagBFP2 | 1 | 200 | 0.031250000 | A6 | 4945 | A | 6 |
| mTagBFP2 | 1 | 200 | 0.015625000 | A7 | 4166 | A | 7 |
| mTagBFP2 | 1 | 200 | 0.007812500 | A8 | 3787 | A | 8 |
| mTagBFP2 | 1 | 200 | 0.003906250 | A9 | 3604 | A | 9 |
| mTagBFP2 | 1 | 200 | 0.001953125 | A10 | 3547 | A | 10 |
| mTagBFP2 | 1 | 200 | 0.000976563 | A11 | 3502 | A | 11 |
| none | 1 | 200 | 0.000000000 | A12 | 3400 | A | 12 |
| mTagBFP2 | 2 | 200 | 1.000000000 | B1 | 50846 | B | 1 |
| mTagBFP2 | 2 | 200 | 0.500000000 | B2 | 29736 | B | 2 |
| mTagBFP2 | 2 | 200 | 0.250000000 | B3 | 15475 | B | 3 |
| mTagBFP2 | 2 | 200 | 0.125000000 | B4 | 9541 | B | 4 |
| mTagBFP2 | 2 | 200 | 0.062500000 | B5 | 6775 | B | 5 |
| mTagBFP2 | 2 | 200 | 0.031250000 | B6 | 5043 | B | 6 |
| mTagBFP2 | 2 | 200 | 0.015625000 | B7 | 4213 | B | 7 |
| mTagBFP2 | 2 | 200 | 0.007812500 | B8 | 3798 | B | 8 |
| mTagBFP2 | 2 | 200 | 0.003906250 | B9 | 3608 | B | 9 |
| mTagBFP2 | 2 | 200 | 0.001953125 | B10 | 3506 | B | 10 |
| mTagBFP2 | 2 | 200 | 0.000976563 | B11 | 3491 | B | 11 |
| none | 2 | 200 | 0.000000000 | B12 | 3593 | B | 12 |
Process data
Process the experimental data using process_plate().
processed_data <- process_plate(
data_csv = "data/example_fluorescence_simple_parsed.csv",
blank_well = c("A12", "B12"),
# timecourse
timecourse = FALSE, ### Note requirement to declare `timecourse = FALSE`
# od
od_name = NULL, ### Note requirement to declare `od_name = NULL`
# fluorescence labels
flu_channels = c("blueblue090"),
flu_channels_rename = c("blueblue"),
# correction
do_quench_correction = FALSE, ### Note that quench correction corrects for cells masking fluorescent signal and is not required
# calibrations
do_calibrate = TRUE,
instr = "spark1",
flu_slugs = c("mTagBFP2"),
flu_gains = c(90),
flu_labels = c("mTagBFP2"),
# conversion factors
od_coeffs_csv = "conversion_factors/od_conversion_factors_assembled.csv",
fluor_coeffs_csv = "conversion_factors/fp_conversion_factors_assembled.csv",
# background autofluorescence subtraction
af_model = NULL, ### Note this is a requirement for `timecourse = FALSE`
outfolder = "experiment_analysis"
)The arguments are described in the ‘Get Started’ vignette. The output CSV is similar.
The following differences apply for process_plate()
where od_name = NULL:
- No requirement for a column labelled
OD600,OD700or similar. - Plots for OD and cell number are not produced.
- Autofluorescence correction does not apply, as autofluorescence is a properly of cells.
- Quench correction does not apply, as this corrects for cellular interference with fluorescence signals.
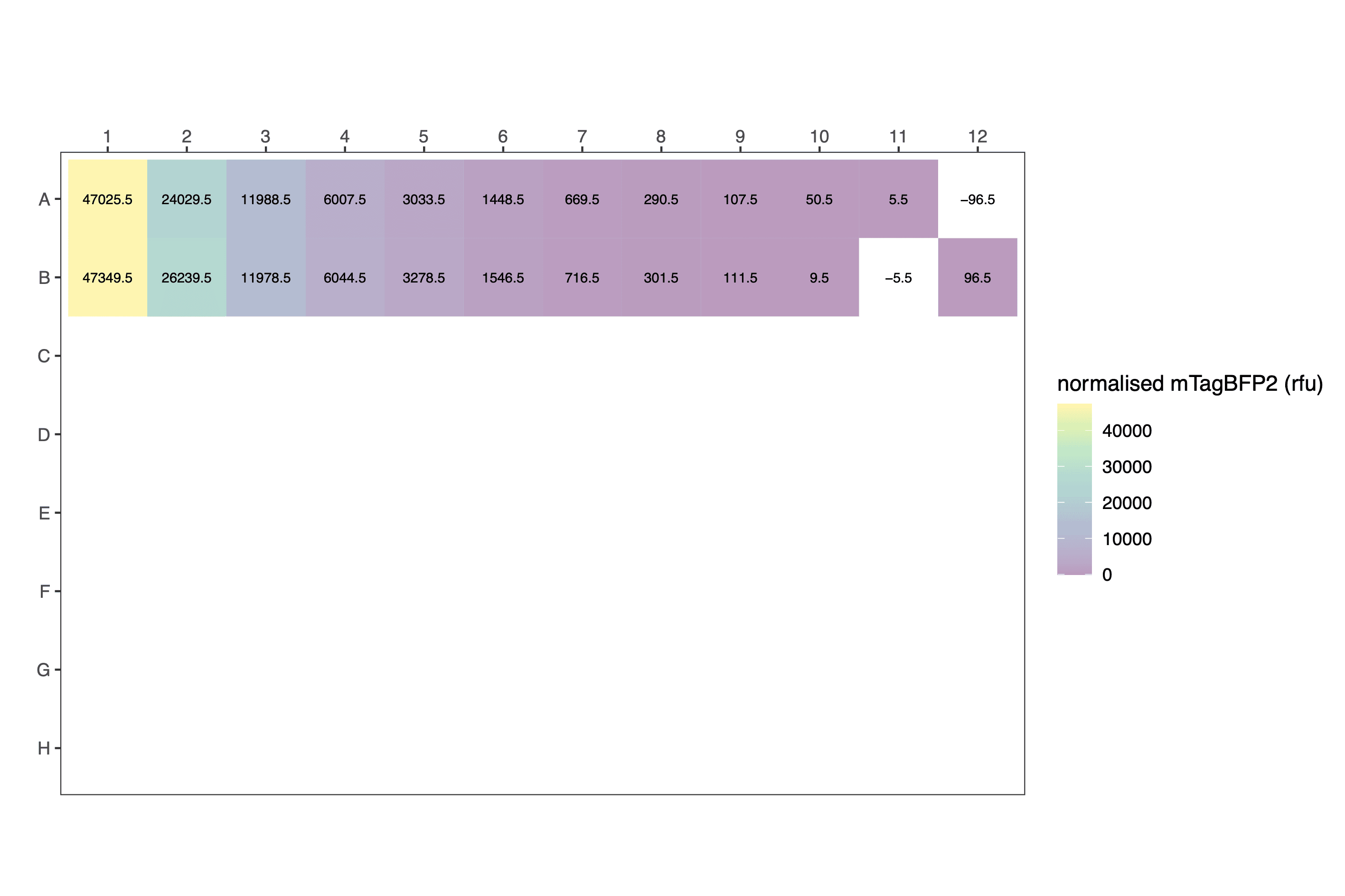
processed_data[1:12,c(1,4,5,6,9,10)] # view a fragment of the dataframe| protein | dilution | well | blueblue | normalised_blueblue | calibrated_mTagBFP2 |
|---|---|---|---|---|---|
| mTagBFP2 | 1.000000000 | A1 | 50522 | 47025.5 | 2.972140e+13 |
| mTagBFP2 | 0.500000000 | A2 | 27526 | 24029.5 | 1.518730e+13 |
| mTagBFP2 | 0.250000000 | A3 | 15485 | 11988.5 | 7.577059e+12 |
| mTagBFP2 | 0.125000000 | A4 | 9504 | 6007.5 | 3.796904e+12 |
| mTagBFP2 | 0.062500000 | A5 | 6530 | 3033.5 | 1.917255e+12 |
| mTagBFP2 | 0.031250000 | A6 | 4945 | 1448.5 | 9.154916e+11 |
| mTagBFP2 | 0.015625000 | A7 | 4166 | 669.5 | 4.231423e+11 |
| mTagBFP2 | 0.007812500 | A8 | 3787 | 290.5 | 1.836039e+11 |
| mTagBFP2 | 0.003906250 | A9 | 3604 | 107.5 | 6.794294e+10 |
| mTagBFP2 | 0.001953125 | A10 | 3547 | 50.5 | 3.191738e+10 |
| mTagBFP2 | 0.000976563 | A11 | 3502 | 5.5 | 3.476150e+09 |
| none | 0.000000000 | A12 | 3400 | -96.5 | -6.099064e+10 |
Output plot formats depend on whether the data is endpoint or timecourse/kinetic data.
Raw:

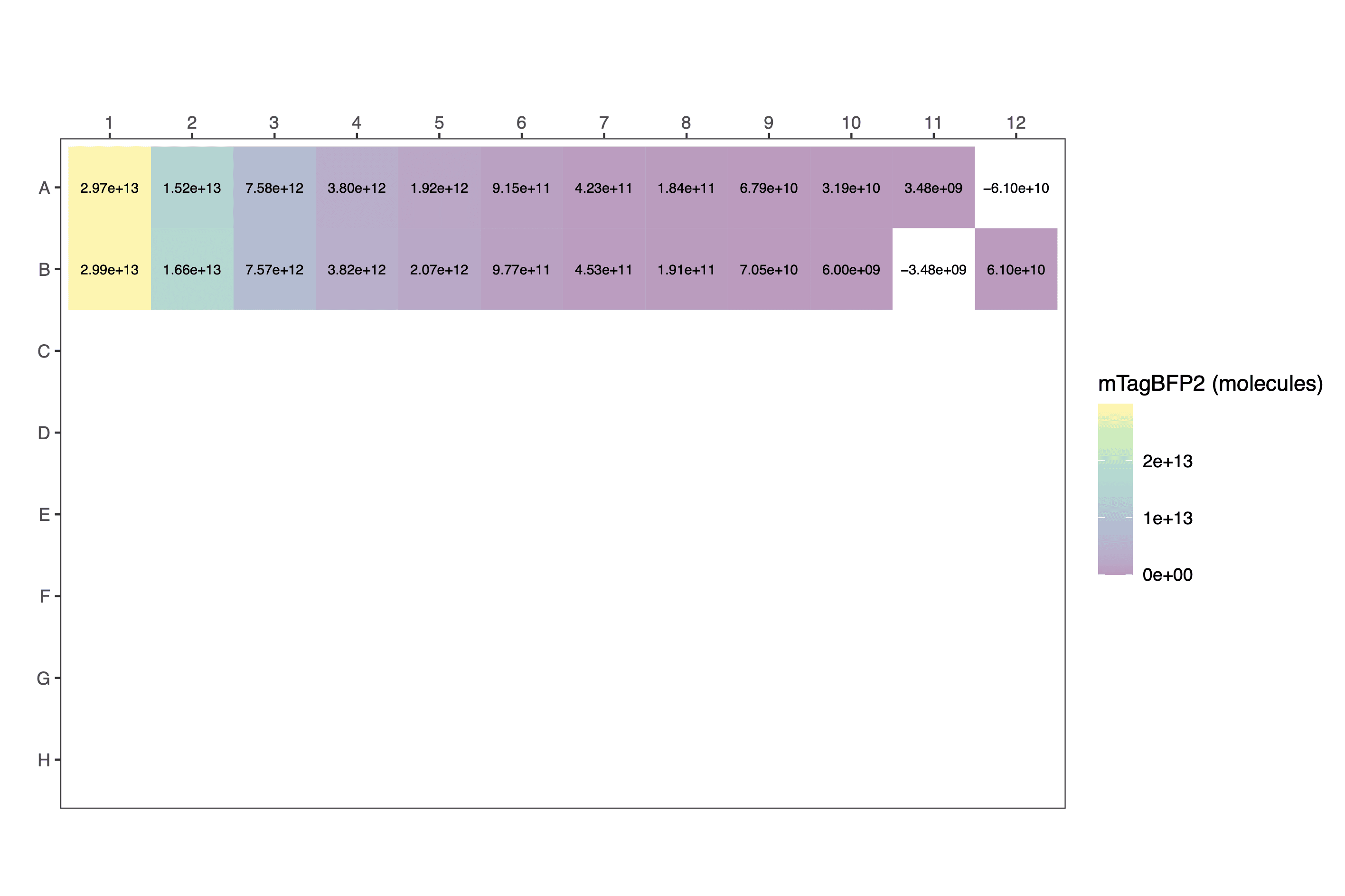
The data can be plotted downstream as a scatter plot or similar.